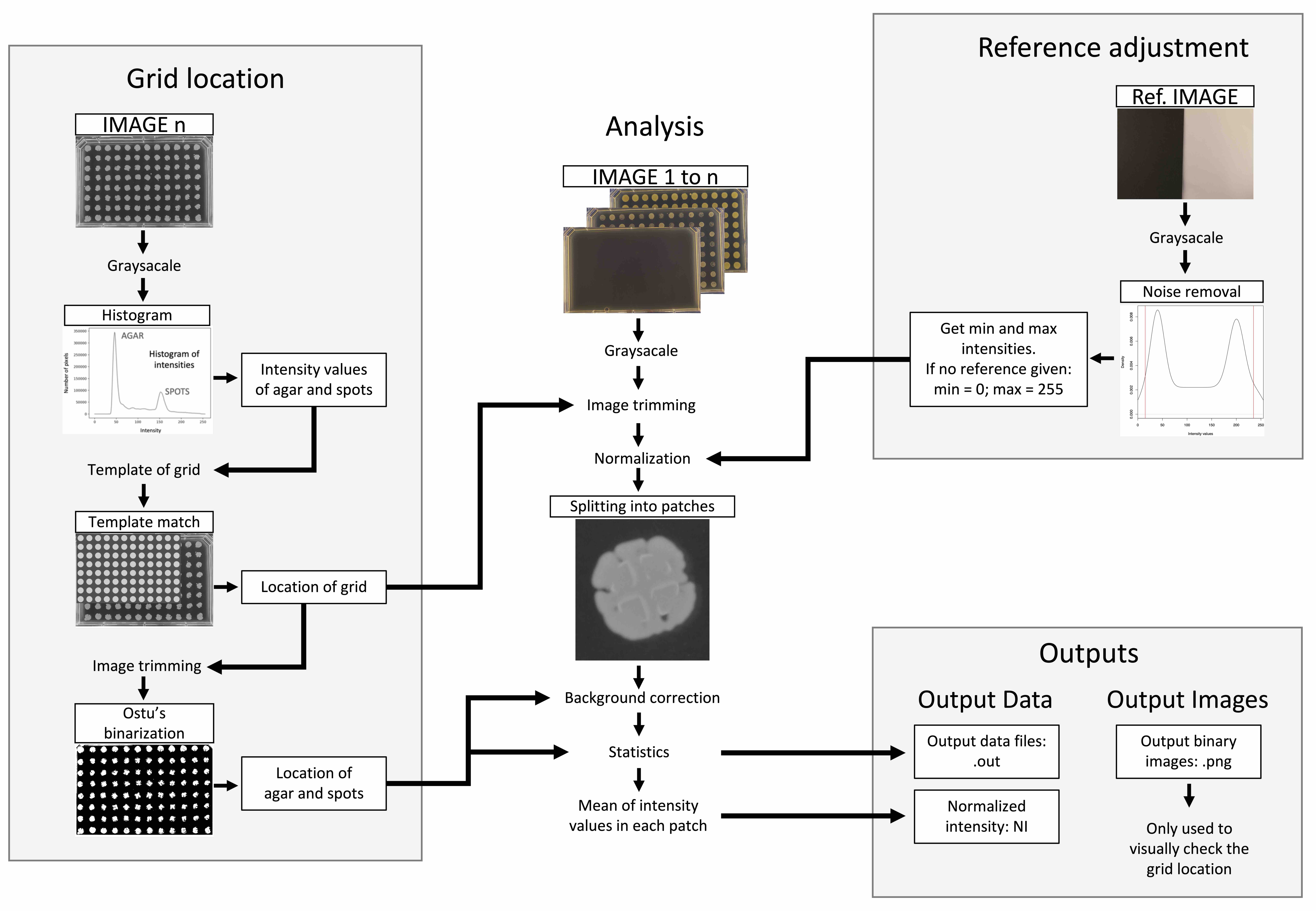
BaColonyzer algorithm
Grid location
BaColonyzer starts the analysis by finding the location of the grid. For that, it imports the grayscale version of the last image in series, which is used to generate a histogram of colors. Colors are represented as intensity values, from 0 to 255.
Since the last image contains well-defined colony spots, the histogram usually shows two high peaks: the highest peak corresponds to the color of the agar, while the second peak shows the color of the colonies (Figure 1). As a quality check, BaColonyzer ensures that the color intensity of the colonies falls between 70% of the maximum color of the picture (upper bound) and twice the color of the agar (lower bound). Otherwise, it sets the color intensity of the colonies to be within this range.
The intensity values of agar and spots are used to create an artificial image of the plate (template), for which users need to input the number of rows and columns of the grid. Resizing and changing the position of this template allows to find the best match with the actual grayscale image (Figure 2). This is achieved by computing the lowest normalized squared difference (The OpenCV Library).
Once the best match is found, BaColonyzer is able to predict the location of the whole plate and to trim those parts of the image that might introduce noise (e.g. the borders). Image trimming, followed by automatic thresholding using Otsu’s binarization (The OpenCV Library), allow to get the position of the colonies and the agar (Figure 3). In order to ensure that pixels belonging to the colonies are never considered as agar, dilation of colony spots is also performed (The OpenCV Library). For that, the dilation kernel is considered to be 5% of the colony area.
Image analysis
Each of the images in series is imported in grayscale and trimmed based on the grid location that has been previously computed. Colony areas are computed using the Otsu's binarization (The OpenCV Library).
By default, color intensities of the image are also normalised (divided by 255) to be in range 0-1. Alternatively, in order to compare different series of time-lapse images, BaColonyzer allows the users to provide a reference picture for the normalization. This must be an image showing a white and black paper next to each other, and must be taken using the same camera settings as the image series (Figure 4). The reference image is imported in grayscale and the color intensities are clipped at 1%-99% quantiles to exclude outiliers or noise. The resulting minimum and maximum intensity values (black and white, respectively) are taken to calibrate the color intensities of images using the expression below:
where NI are the new intensity values of each image, OI are the original intensity values, min_{ref} is the intensity of the black color, and max_{ref} is the intensity of the white color.
Next, in order to ensure an accurate analysis of each colony, BaColonyzer divides the agar plate into smaller patches that contain one colony spot. By default, the tool adjusts the intensity values of each patch by subtracting the mean intensity of the agar in this patch, thus correcting for color differences within and between images (Figure 5).
At the end, BaColonyzer provides a big table with statistics of each colony, including normalised intensities (NI) of each patch, colony area, colony mean, colony variance, background mean, and background variance. Normalized intensity values of each patch (NI) are computed as the sum of all intensity values of the patch, divided by the number of pixels. Since all patches have exactly the same number of pixels, bigger colonies will result in higher normalized intensities, allowing to account also for the colony size. Results are stored as Output Data. Furthermore, in order to visually check that the grid location was achieved properly, BaColonyzer provides some Output Images. These are binary images resulting from an Otsu’s Binarization (The OpenCV Library), which is performed after the trimming step to let the users check whether the grid location was successful.
Flowchart
A flowchart showing the algorithm of BaColonyzer is found below:
Enjoy using BaColonyzer!